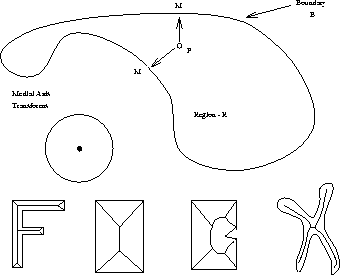
Figure 5: Definition and examples of medial axis transforms


The medial axis transform is a skeletal representation of regions which is applied commonly to binary imagery, but is extensible to more complex data. It is defined as following, with reference to Figure 5, below.

Figure 5: Definition and examples of medial axis transforms
For each point P in region R, find M the nearest neighbour on boundary B. The nearest neighbour is the point such that the distance P to M is minimum. If P has more than one nearest neighbour, then P is a skeletal point The union of the skeletal points is the Medial Axis Transform. If the straight sections are then linked this is the Medial Line Transform.
The MAT is a succinct representation with low storage requirements -- the original image may be reconstructed from the MAT. Features of the MAT can be used for recognition, e.g. the number of nodes, branches etc. Notably, it is applied in the medical field to chromosome identification.
However it has several disadvantages. While fairly reliable with thin objects eg the letter F or chromosomes, it is less good with thicker objects. It is very susceptible to minor flaws in the boundary, as illustrated by the rectangular shape in Figure 5.
Algorithm for MAT
Compute the distance function, i.e. Label each regional pixel with a number denoting the distance to the boundary (This can apply to either 4- or 8- connected images.) Mark all local maxima
Equivalently:
Expand a circle from each pixel within the region. Note when the circle touches the boundary If it touches at more than one point, then the centre is on the MAT.
Refinements
Link all local maxima by a minimum distance link, X. Thin by permitting each pixel only one neighbour Remove noise spurs of unit length.

Figure 6: Thinning and computing the MAT



[ Run-length coding |
Quad trees ]
Comments to: Sarah Price at ICBL.