SIGNAL PROJECT
Signal transduction pathways are biochemical pathways which allow cells to sense a stimulus and communicate a signal to the nucleus, which then makes a suitable response. They are complicated signalling processes with built-in feedback mechanisms.
Signalling pathways are embedded in larger networks and are involved in important processes such as proliferation, cell growth, movement, cell communication, and programmed cell death (apoptosis). Malfunction results in a large number of diseases including cancer, diabetes and many others.
Despite enormous experimental advances in recent years there is still an absence of good, predictive pathway models which can guide experimentation and drug development. To date, models either encode static aspects such as which proteins have the potential to interact, or provide simulations of system dynamics using ordinary differential equations.
Background
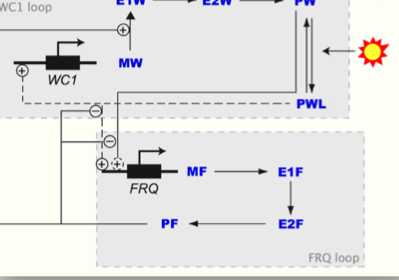
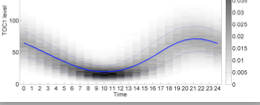
We are developing a novel approach to analytic pathway modelling based on our experience of modelling concurrent computing systems.
We model pathways using stochastic process algebras which denote continuous time Markov chains thus affording new quantitative analysis and new ways to reason about incomplete behaviour.