Factorial ANOVA
In the same way as DV ~ IV is R formula for linear
regression or one way ANOVA, the formula for two-way ANOVA is DV ~ IV1
* IV2 or DV ~ IV1 + IV2 + IV1
: IV2
Especially the latter one illustrates that we're looking at two predictors and the interaction between those.
Now, we'll apply it to ToothGrowth dataset provided in R.
First, load it:
> attach(ToothGrowth)
Look
at the dataset:
> head(ToothGrowth)
Dose
is treated as numerical variable, instead we want it to be categorical, let's code
the factors
> ToothGrowth$dose = factor(ToothGrowth$dose, levels=c(0.5,1.0,2.0), labels=c("low","med","high"))
See what changed:
> head(ToothGrowth)
Start with some descriptive statistics:
> boxplot(len ~ supp
* dose, data=ToothGrowth,
+ ylab="Tooth Length", main="Boxplots of Tooth
Growth Data")
You might also check histograms of each
factor combination to decide if the mean is an appropriate summary statistic.
If this looks okay a useful way to summarise the overall pattern of two-way
data is an interaction plot:
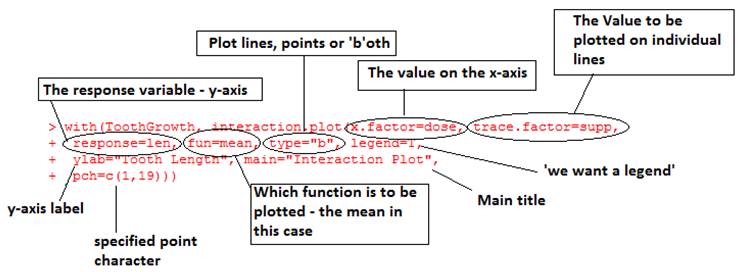
> interaction.plot(x.factor=dose, trace.factor=supp, response=len, fun=mean, type=”b”)
And test the assumption that
variances are equal:
> bartlett.test(len ~ supp
* dose, data=ToothGrowth)
Now, let's look at the model and the
summary:
> model=lm(len~supp*dose)
> summary(model)
also, look at summary.lm() output to see all coefficients.
Same as for one way ANOVA we can also call aov() function:
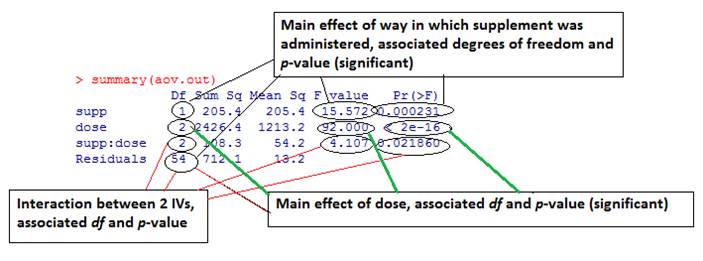
> aov.out = aov(len ~ supp * dose, data=ToothGrowth) > summary(aov.out)This allows us to look at the results of planned contrasts: > summary.lm(aov.out) Note that the interaction is significant so we should be careful about interpreting the main effects. We can run post-hoc analysis between all 3*2=6 conditions: > TukeyHSD(aov.out) Or we can restrict it to one predictor (dose): > TukeyHSD(aov.out, which=c("dose")) We can test
for the effect without the interaction:
> model2=lm(len~supp+dose)
Or could do
with interaction but without one main effect
> model3=lm(len~supp+supp:dose)
And could
compare whether one model explains significantly more than the other:
> anova(model,model2)
In fact the
ANOVA test between models assumes that the simpler model is a strict subset of
the other. Is that true in this case?