Generative models are now used to find designs for new molecules for medicines. These models can produce thousands of molecules targeted to bind with a particular protein, but most such designs do not make suitable medicines. A common problem with generated designs is that they can bind with off-target proteins where we do not want it to bind. The side effects due to such off-target binding can be serious and even lethal. Thus the molecules have to be screened through various tests to filter out the ones with off-target effects. We need models that can directly avoid off-target effects during the generative design.
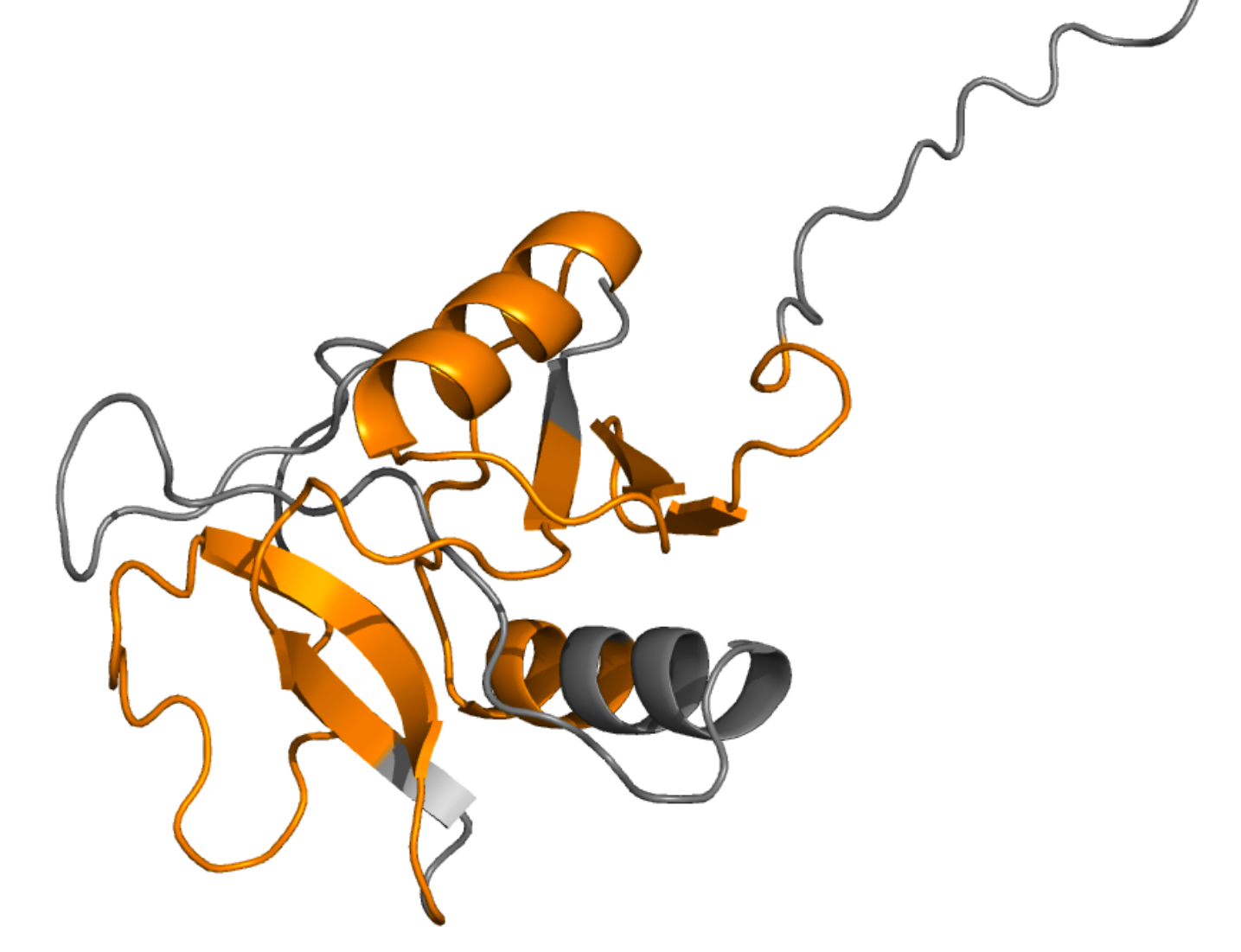
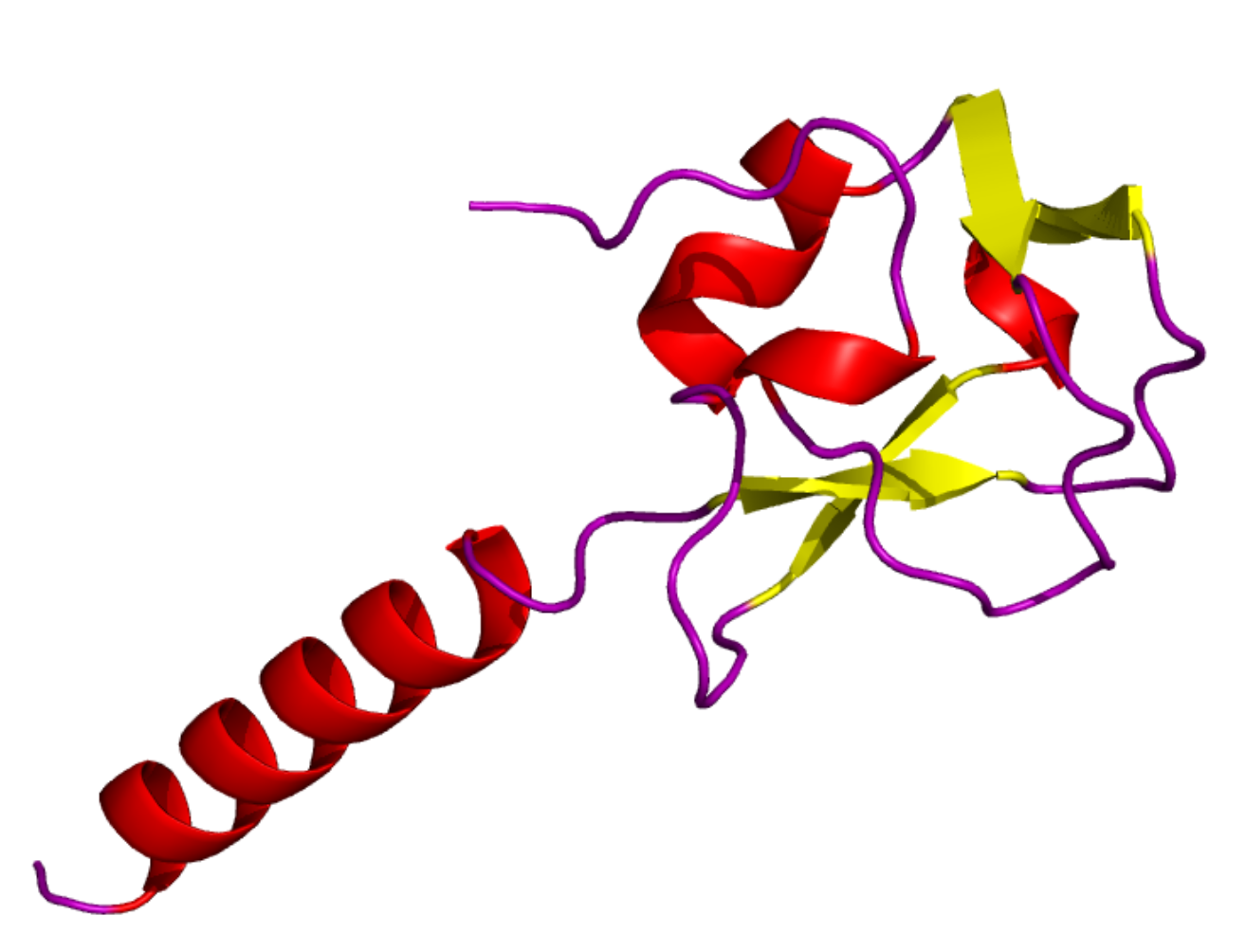
The challenge in drug and protein analysis comes from their complex shapes. Above you see pictures of two proteins from one of our projects (thanks to Chris Crampton for the images). A drug-protein interaction is a question of the drug fitting with a part of the protein. Generative models operate by searching a space of possible drug shapes to find such a fit. The plan in this project is to dive deeper into the geometry of molecule shapes and guide the generative process to avoid avoid fitting with off-target protein shapes. We will combine generative AI with computational geometry, topology, and various kinds of shape analysis and statistical techniques to design new generative models.
The student will have the freedom to explore and develop the project according to their preferences. They will have the opportunity to learn a range of ides and techniques in generative AI, topological data analysis, and various other areas of machine learning, mathematics and biology that interest them. The project is part of the CDT in AI for Biomedical Engineering, and includes option for an Internship. It will be a collaboration with Dr. Chris Wood at School of Biology in Edinburgh, Oxford Drug Design — a drug designing company, and many other people.
Application deadline is in January 2026.