Research - this page is getting a long needed refresh, please bear with me
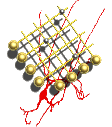
Synapse Protein Networks
![[G2C LOGO]](g2c.png)
Synapses are the fundamental unit of computation in the brain playing key roles in information processing, behaviour and disease. They not only transmit information between cells but they also detect patterns of neural activity and process this information by activating intracellular biochemical signalling pathways, which subsequently changes the properties of the neuron.
We started work in this area and a long term collaboration with Prof Seth Grant as part of the Wellcome Trust funded Genes2Cognition programme. In a series of projects since G2C including the EU supported EuroSpin, SynSYS and Human Brain Project consortia we have developed a series of methodological apporaches to analysing proteomic networks at the synapse (and elsewhere). In research lead by Dr Oksana Sorokina in the group we also maintain a synapse proteomic datatabase and suite of tools.
Actual Analytics
Actual Analytics: Modern molecular science can study entire genomes in a single experiment (10s of thousands of genes) yet behavioural research is still largely rooted in a human observer watching the animals behave, especially within social or interactive environments. Our studies into individual annotators/experts show that individual interpretation of behavioural events can have a huge impact in experimental studies. We have been developing a range of tools that automate behavioural analysis and data capture. These tools are essential to bridge the gap between the level of analysis we can perform in large protein complexes to their phenotypes in integrated studies. Further, the use of computer vision and tracking algorithms can capture information that is extremely hard to obtain using any other method. These techniques are also providing new insights into behaviour. Using techniques from machine learning we have developed a new approach to these problems where our system learns directly from experts how to recognise the behaviour under investigation. Studies in a range of animal behaviours demonstrate that we can effectively mimic a human expert.