| The Bio-PEPA Eclipse Plug-in User Manual |
In this example (see Table 3), an enzyme E combines with a substrate S to form a compound E:S. This compound might degrade releasing the enzyme and the substrate or it might convert the substrate into product P, releasing the enzyme (In the Bio-PEPA Eclipse Plug-in editor the two parallel diagonal lines (//) denote comments). The Outline View of the model is shown in Figure 74. According to the species components definition:
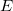 — is a reactant (
— is a reactant (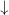 , <<) in reaction
, <<) in reaction 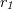 and a product (
and a product ( , >>) in reactions
, >>) in reactions 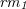 and
and 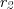
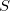 — is a reactant (
— is a reactant (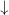 , <<) in reaction
, <<) in reaction 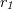 and a product (
and a product ( , >>) in reaction
, >>) in reaction 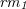
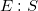 — is a product (
— is a product (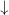 , <<) in reaction
, <<) in reaction 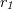 and a reactant (
and a reactant ( , >>) in reactions
, >>) in reactions 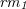 and
and 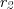
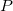 — is a product in reaction
— is a product in reaction 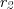
The Bio-PEPA syntax |
//The Bio-PEPA plugin syntax |
Parameter Definitions: |
//Parameter Definitions |
|
k1 = 1.0; |
|
km1 = 0.1; |
|
k2 = 0.01; |
|
|
Functional Rates |
//Functional Rates |
|
r1 = [ k1 * E * S ]; |
|
rm1 = [ km1 * E:S ]; |
|
r2 = [ k2 * E:S ]; |
|
|
Species Components |
//Species Components |
|
E = r1 << + rm1 >> + r2 >> ; |
|
S = r1 << + rm1 >> ; |
|
E:S = r1 >> + rm1 << + r2 << ; |
|
P = r2 >> ; |
|
|
Model Component |
//Model Component |
|
E[40] <*> S[30] <*> E:S[0] <*> P[0] |
![\includegraphics[scale=0.5]{screenshots/screenshots/mmoutline}](images/img-0177.png)