Most region-based methods are simple, easy to implement, and execute
fairly quickly. They tend to perform well in tasks such as area and
volume calculations, and classification of easily distinguishable
tissues. However, they do have certain drawbacks that can limit their
usefulness. First, they provide no structural information about the
regions they classify, because it is generally difficult to extract
geometric information from a collections of pixels. Second, they can
be confused by image anomalies such as false bridges between close,
but separate regions, or by occlusions such as one region blocking the
view of another region (see
Figure ![]() ). More sophisticated
region-based methods include Fourier pattern
analysis [77] and neural networks models [48].
In Fourier analysis, regions are decomposed into a regular grid
pattern and an n-dimensional Fourier transform is applied to each
subregion. The frequency and intensity distribution of transformed
data can be compared with the expected distribution from training
samples. Neural networks are similar to statistical image processing
methods with the exception of the presence of hidden nodes that can
regroup and transform the input data patterns. Although neural
networks themselves suffer from many drawbacks, such as sensitivity to
geometric scale, orientation, and intensity distribution, they have
nonetheless been used with varying degrees of success in thousands of
applications.
). More sophisticated
region-based methods include Fourier pattern
analysis [77] and neural networks models [48].
In Fourier analysis, regions are decomposed into a regular grid
pattern and an n-dimensional Fourier transform is applied to each
subregion. The frequency and intensity distribution of transformed
data can be compared with the expected distribution from training
samples. Neural networks are similar to statistical image processing
methods with the exception of the presence of hidden nodes that can
regroup and transform the input data patterns. Although neural
networks themselves suffer from many drawbacks, such as sensitivity to
geometric scale, orientation, and intensity distribution, they have
nonetheless been used with varying degrees of success in thousands of
applications.
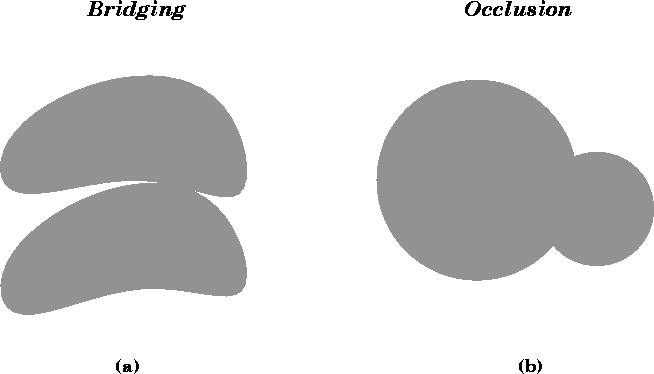
Figure: Illustration of two potential limitations of region-based
classification methods. (a) Due to the limited spatial resolution
of CT and MR images, bridging errors can occur between distinct
anatomical structures. (b) Occlusion errors occur when one region
overlaps another region and partially blocks the perceived
boundary of one or both of these regions.