| The Bio-PEPA Eclipse Plug-in User Manual |
One of the most intuitive and direct kinds of analysis which can be applied to a Bio-PEPA model is to simulate the model and plot the quantities of the chemical species in the model as a function of time. (Such a plot is called a time series.) The relative amounts of the chemical species in the model will change over time because the reactions in the model change the proportions of species.
When we invoke the Time-series Analysis option from the Bio-PEPA menu we are presented with a wizard which guides us through the process of selecting a simulation method and plotting results (see Figure 18). In our simple example, with only one species 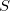 which decays there is only one option here, to plot
which decays there is only one option here, to plot 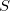 .
.
For models with many species you can narrow the list of species by typing a species name (or part of a species name) in the search box in the upper right hand corner. The list of species will narrow to include only those species which contain this search string in their name.
If we do not select any species to plot then we will not be able to proceed (see Figure 19).
![\includegraphics[scale=0.5]{screenshots/dialogues/timeseriesanalysiswizard}](images/img-0041.png)
![\includegraphics[scale=0.5]{screenshots/dialogues/timeseriesblocked}](images/img-0042.png)
The next step in generating a time series is to choose the kind of simulation which is to be done. Different types of simulators are available in the Bio-PEPA Eclipse Plug-in, including:
continuous, deterministic simulators which convert your Bio-PEPA model into a system of Ordinary Differential Equations (ODEs) which are evaluated using numerical integration; and
discrete, stochastic simulators which convert your Bio-PEPA model into a Monte Carlo Markov Chain (MCMC) problem which is evaluated using exact or approximate stochastic simulation algorithms such as Gillespie’s Direct Method and Gillespie’s  -leap algorithm.
-leap algorithm.
We will describe each of these in turn.