| The Bio-PEPA Eclipse Plug-in User Manual |
As an example, we will use a model of a general genetic network with a negative feedback through dimmers (see Figure 1).
![\includegraphics[scale=0.9]{screenshots/screenshots/GeneticNetworkPng}](images/img-0178.png)
The model, which is shown in Table 4, is composed of three biological entities that interact with each other through five reactions (of which one is reversible). The biological entities are the mRNA molecule (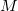 ), the protein in monomer form (
), the protein in monomer form (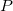 ) and the protein in dimeric form (
) and the protein in dimeric form ( ). The first reaction (1) is the transcription of the mRNA (
). The first reaction (1) is the transcription of the mRNA (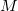 ) from the genes/DNA (not considered explicitly). The protein
) from the genes/DNA (not considered explicitly). The protein 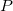 in the dimer form (
in the dimer form (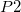 ), which is the final result of the network, has an inhibitory effect (
), which is the final result of the network, has an inhibitory effect ( , (-)) on this process. The second reaction (2) is the translation of the protein
, (-)) on this process. The second reaction (2) is the translation of the protein 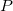 from
from 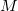 , in which
, in which 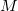 acts as an activator (
acts as an activator (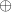 , (+)). The other two reactions represent the degradation of
, (+)). The other two reactions represent the degradation of 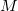 (3) and the degradation of
(3) and the degradation of 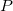 (4). Finally there is the dimerization of
(4). Finally there is the dimerization of 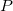 and its inverse process (5, 5i). All the reactions are described by mass-action kinetics with the exception of the first reaction, which has a Michaelis-Menten kinetics [11] (see section A.4 for more information on mass-action and Michaelis-Menten kinetics) .
and its inverse process (5, 5i). All the reactions are described by mass-action kinetics with the exception of the first reaction, which has a Michaelis-Menten kinetics [11] (see section A.4 for more information on mass-action and Michaelis-Menten kinetics) .
The Outline View of the model can be seen in Figure 76.
The Bio-PEPA syntax |
//The Bio-PEPA plugin syntax |
Locations: |
//Locations |
|
location v_cell: size = 1; |
|
|
Parameter Definitions |
//Parameter Definitions |
|
KM = 356; |
|
v = 2.19; |
|
k2 = 0.043; |
|
k3 = 0.039; |
|
k4 = 0.0007; |
|
k5 = 0.025; |
|
k5i = 0.5; |
|
|
Species Attributes |
|
|
|
|
|
|
|
|
|
Functional Rates |
//Functional Rates |
|
a1 = [v/(KM+P2)]; |
|
a2 = [fMA(k2)]; |
|
a3 = [fMA(k3)]; |
|
a4 = [fMA(k4)]; |
|
a5 = [fMA(k5)]; |
|
a5i = [fMA(k5i)]; |
|
|
Species Components |
//Species Components |
|
M = a1 >> + a2 (+) + a3 << ; |
|
P = a2 >> + a4 << + (a5,2) << + (a5i,2) >>; |
|
P2 = a1 (-) + a5 >> + a5i <<; |
|
|
Model Component |
//Model Component |
|
M[0] <a2> P[0] <a5, a5i> P2[0] |
![\includegraphics[scale=0.5]{screenshots/screenshots/gennetworkoutline}](images/img-0202.png)