| The Bio-PEPA Eclipse Plug-in User Manual |
The species component definition, lists all the reactions a particular species is allowed to participate in and in what role. The list of actions are separated by the choice operator 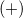 . The concrete syntax, as accepted by the plug-in, is as follows [14]:
. The concrete syntax, as accepted by the plug-in, is as follows [14]:
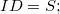
where 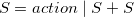
and where an 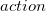 is defined as:
is defined as:
![$ \begin{array}{r} action \; = \; (rateID, \; stoichiometry) \; op \; speciesID; \; | \; rateID \; op \; speciesID; \; |\; \\ (\; rateID\; [\; locationID \; op’ \; locationID\; ], \; stoichiometry\; ) \; (.) \; ID; \; |\; \\ rateID\; [\; locationID \; op’ \; locationID\; ] \; (.) \; ID; \end{array} $](images/img-0140.png)
with
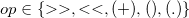
and
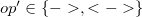 .
. The first  refers to a species, but in this particular definition is labelled as an
refers to a species, but in this particular definition is labelled as an  to make it distinct from the the other species identifiers expected within a single species component definition. This identifier must take the first
to make it distinct from the the other species identifiers expected within a single species component definition. This identifier must take the first 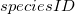 form of just the species name with no location. Thus, each global species can have at most one definition, with the
form of just the species name with no location. Thus, each global species can have at most one definition, with the 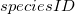 used to allow control over where a particular action is permissible. In the case of non-transport definitions, i.e. the first two in the action definition, the
used to allow control over where a particular action is permissible. In the case of non-transport definitions, i.e. the first two in the action definition, the 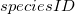 can take any of the previously defined forms. Thus a single reaction can be defined as occurring in the global sense for the species, in a single location, or in a subset of the locations where the species is present [14]. If a reaction occurs in the global sense for the species, then the
can take any of the previously defined forms. Thus a single reaction can be defined as occurring in the global sense for the species, in a single location, or in a subset of the locations where the species is present [14]. If a reaction occurs in the global sense for the species, then the 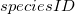 identifier may be ommited. For example, in the species components definitions of the a-b-c.biopepa model (see Figure 42), the
identifier may be ommited. For example, in the species components definitions of the a-b-c.biopepa model (see Figure 42), the 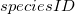 identifier after the operation symbol has been ommited in all of the reactions, as they occur in the global sense :
identifier after the operation symbol has been ommited in all of the reactions, as they occur in the global sense :
This is equivalent to:
The identifiers labelled 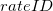 must refer to a previously defined functional rate. Additionally,
must refer to a previously defined functional rate. Additionally, 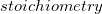 is the stoichiometric coefficient for the species in this reaction(
is the stoichiometric coefficient for the species in this reaction(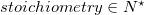 ). If a statement does not specify a stoichiometric coefficient, the default value of one is used. The definition for
). If a statement does not specify a stoichiometric coefficient, the default value of one is used. The definition for 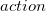 shows the syntax for including the stoichiometric coefficient and the abbreviated form for when the stoichiometry is one, these being the first two forms shown in the action definition above [14]. For example, in the species components definitions of the a-b-c.biopepa model (see Figure 42), the stoichiometric coefficient has been omitted, since it has the value of 1 in all cases:
shows the syntax for including the stoichiometric coefficient and the abbreviated form for when the stoichiometry is one, these being the first two forms shown in the action definition above [14]. For example, in the species components definitions of the a-b-c.biopepa model (see Figure 42), the stoichiometric coefficient has been omitted, since it has the value of 1 in all cases:
This is equivalent to:
And also equivalent to:
Transportation represents the movement of a single species between two adjacent locations within the model, as described in [10]. In the defined syntax above (third and fourth action definitions) it requires two 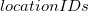 , the first acting as the source and the second as the target and must refer to locations that this particular species resides in. As the locations are embedded within the transportation action, the identifier is simply the species name. The general modifier operator ((.)) is used for transportation as, while the levels of the species in the two locations change, the overall amount does not. Transportation can either be in a single direction or in both directions (unidirectional or bidirectional transportation respectively) [14]. For an example of the unidirectional and the bidirectional transportation see section A.9 and section A.10, respectively.
, the first acting as the source and the second as the target and must refer to locations that this particular species resides in. As the locations are embedded within the transportation action, the identifier is simply the species name. The general modifier operator ((.)) is used for transportation as, while the levels of the species in the two locations change, the overall amount does not. Transportation can either be in a single direction or in both directions (unidirectional or bidirectional transportation respectively) [14]. For an example of the unidirectional and the bidirectional transportation see section A.9 and section A.10, respectively.
For the Bio-PEPA Eclipse Plug-in user’s convenience, many of the original Bio-PEPA mathematical symbols have been replaced with textual representations that are available on every keyboard. Table 1 contains the Bio-PEPA symbols and their ASCII representations in the Bio-PEPA Eclipse Plug-in [14]:
Behaviour |
BioPEPA symbol |
ASCII representation |
reactant |
|
<< |
product |
|
>> |
activator |
|
(+) |
inhibitor |
|
(-) |
modifier |
|
(.) |
unidirectional transportation |
|
-> |
bidirectional transportation |
|
<-> |