SFARI Genes and How to Find Them
We recently published an article in Scientific Reports based on work by Magdalena that studied the relationship between gene expression profiles of patients with Autism Spectrum Disorders and their status as genes already identified as having a role in the development of ASDs. Put simply, do these genes have expression features that we can learn so that we can build models to identify potential novel ASD causative genes? In this paper we show that, despite the signal being quite weak, there is information that we can leverage from high throughput gene expression studies to give us indications of new genes for ASD. Interestingly, in order to find this signal Magdalena had to integrate gene exrpression profiles across the entire expression landscape by creating weighted gene correlation networks. Only by sharing the information across the whole system was it possible to develop an informative statistical model for candidate prioritisation.
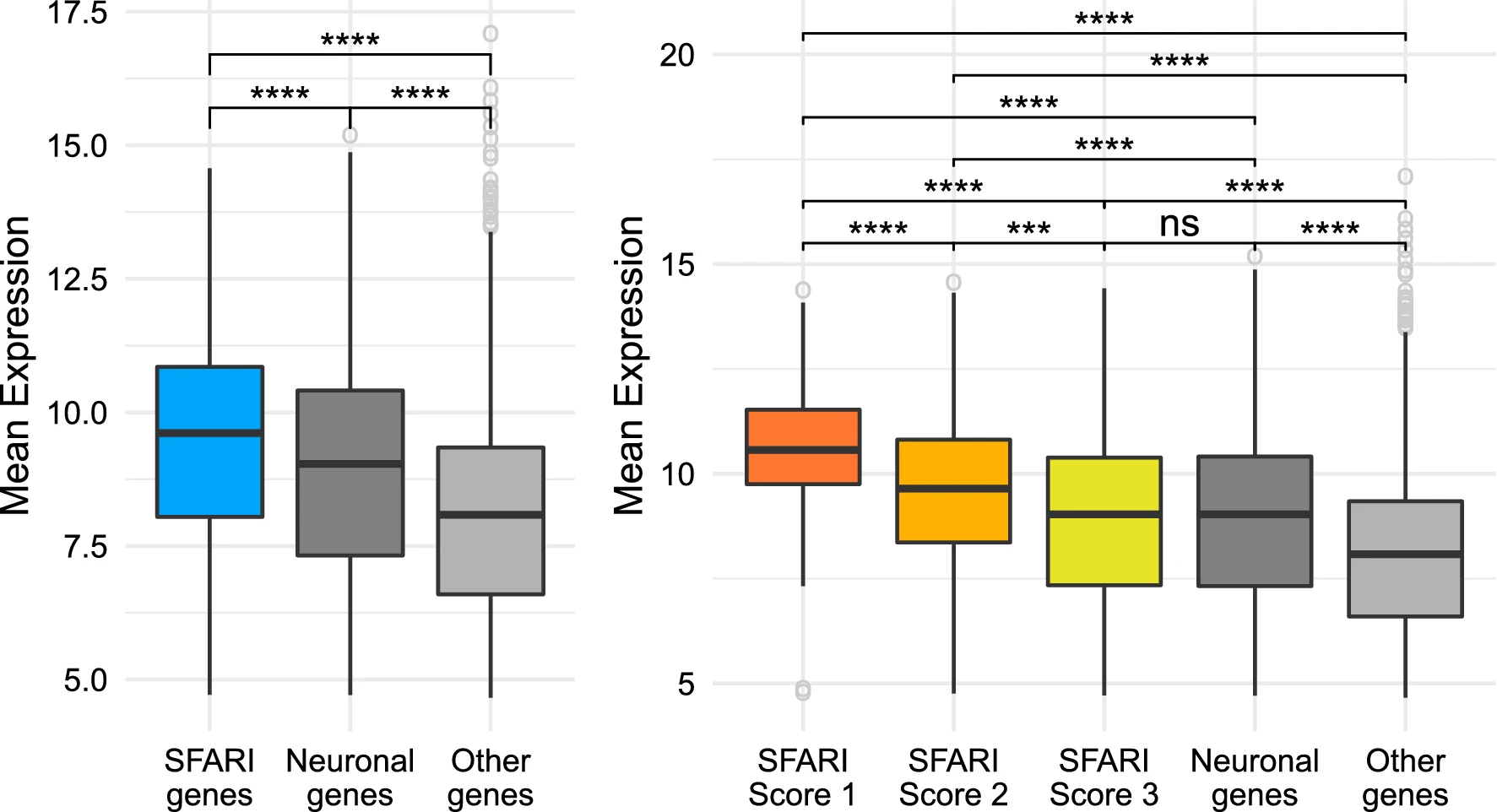
SFARI genes have higher levels of expression than other genes. Comparison between the SFARI genes, genes with neuronal annotations and with the rest of the genes in the dataset. The brackets at the top indicate pairwise comparisons, using a Welch t-test to study wether the differences in level of expression between groups are statistically significant, and the asterisks indicate the magnitude of the corrected p value of each test: ns = p value ≥ 0.05, *p value < 0.5, \*\*p value < 0.01, \*\*\*p value < 0.001, and \*\*\*\*p value < 0.0001. (A) SFARI genes. (B) SFARI Scores. Outlier genes are represented individually as open circles. The t-tests use all the points in each group, including outliers.
The paper contains a lot of detail about the pitfalls to avoid and important considerations to address related to bias, confounding variables, and other sources of technical error that have a large impact on how gene expression data should be analysed and interpretted. These are very often not considered in published studies so we believe that this makes the paper an even more interesting contribution to the field; even beyond ASD research.
Whilst these observations were replicated in 3 independent data sets, what was also clear is that, as you might expect, data is all. The heterogeneity of samples in terms of quality, brain region, age, and likely most importantly ASD severity are critical. Due to the nature of the data available we didn’t have the statistical power to separate out information by brain region and crucial patitent meta-data was absent preventing us from looking at the impact of ASD severity in these relationships.
If this sounds interesting please do check out Magdalena’s paper here.