Network based approach to identify sub-populations within Parkinson's Disease
Barry Ryan (e-mail) is a PhD student from the CDT in Biomedical Artificial Intelligence working in the group on network based approaches in biomedicine, specically aiming to integrate multi-modal data to improve our understanding of neurological disease.
Abstract
Precision medicine is a term coined to describe the movement of medicine towards a personalised preventative approach compared to current reactionary practices. Diseases, individuals and environments are diverse, however current medical practice looks to group together common symptoms under a single disease. Precision medicine seeks to move away from this and identify common causes and manifestations of diseases which pre-date symptoms thus, facilitating preventative treatments. Central to the development of precision medicine is therefore the integration of genetic, environmental and lifestyle data for accurate disease classification.
Networks are all around us. A computer network is a set of computers sharing resources located on network nodes. A social network, such as Facebook or Twitter, is a set of humans sharing information such as photos and posts with each other. A patient similarity network (PSN) is a set of individuals sharing medical information in an attempt to identify commonalities or similarities within a disease. PSN's can be used to integrate multi-modal data sources to classify patients in an interpretable manner (Pai and Bader 2018). In theory, a PSN could be used to; identify novel genetic mutations, improve the understanding of the pathology of disease, improve candidate selection in clinical trials and identify individuals who are at a higher than average risk of a disease.
Parkinson’s disease (PD) is a heterogeneous disease with multiple causes and manifestations many of which remain unknown (Dextera and Jenner 2013). Currently therapies are aimed at relieving symptoms of PD rather than addressing the underlying causes. While genetic causes have been shown to account for 30% of cases, much about the pathology of the disease remains unknown (Klein and Westenberger 2012). Furthermore, common biological pathways between genetic and idiopathic (no known genetic cause) PD only converge late in the disease course. Other previous research has found associations with genetic, environmental and lifestyle factors and PD. For these reasons, PD is a perfect testing ground on which to test a network-based approach for precision medicine.
Research Plan
First Year
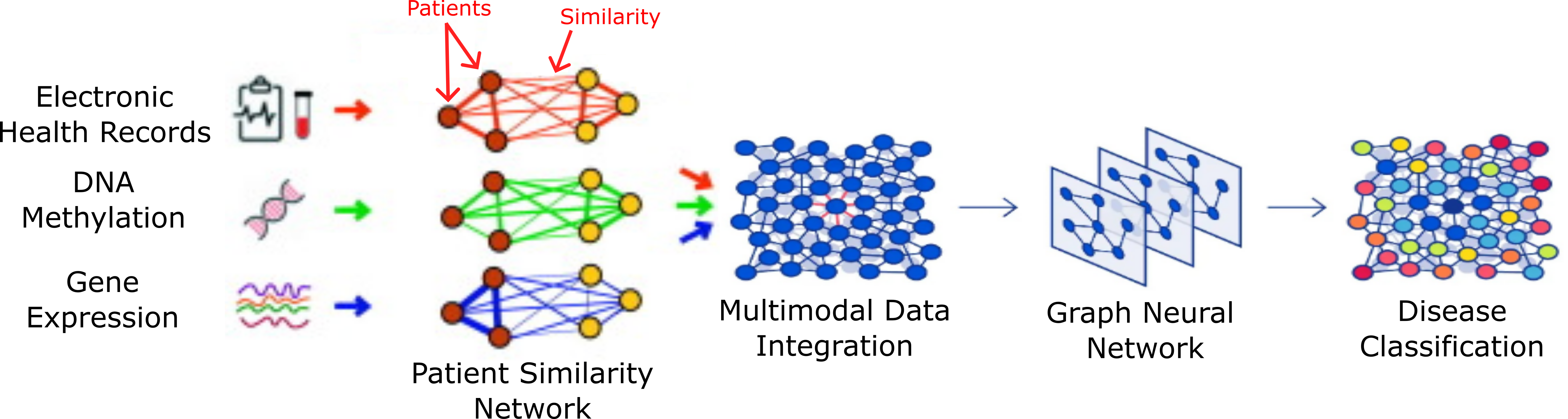
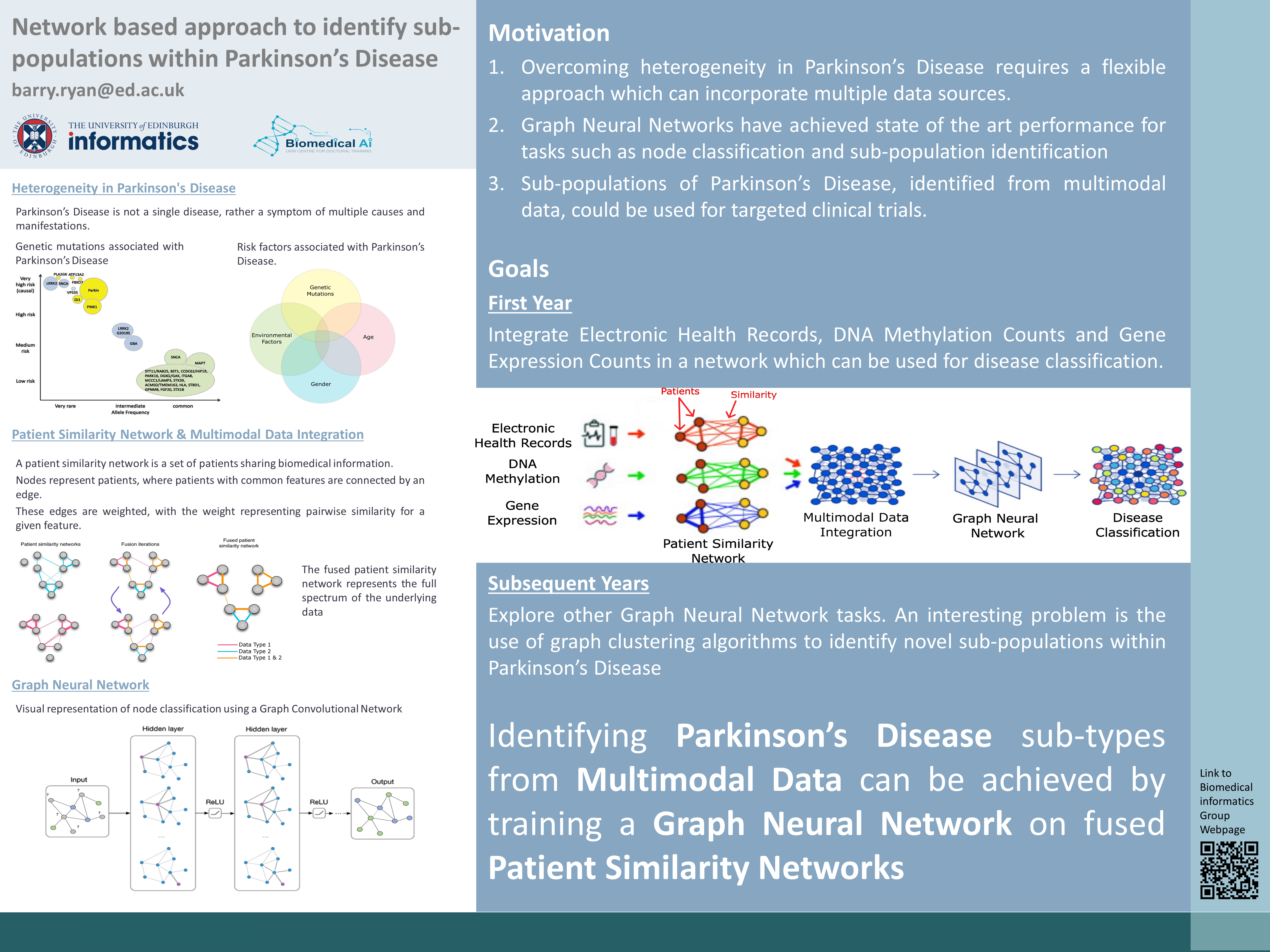
The first stage of the research plan looks to integrate Electronic Health Records with Gene Expression and DNA Methylation datasets. The data is obtained from the Parkinson's Progressive Marker Initiative (PPMI) study (see below). The goal of this stage is to develop a pipeline for multimodal data integration using patient similarity networks (PSN). From the integrated network a Graph Neural Network (GNN) can be trained. GNN's are a particular type of neural network designed to handle unstructured graph data. Common GNN tasks include edge prediction, node classification, community detection and graph level prediction. Node classification is one of the simpler GNN prediction tasks and corresponds to disease classification, hence this was chosen as the initial research test goal. It is expected that integration of multiple data sources will provide better classification in Parkinson's Disease individuals compared to current state of the art methods.
Subsequent Years
Future years will look to integrate more data types provided initial success. There is also the possibility to implement other Graph Neural Network (GNN) tasks. A potentially very useful and interesting application is the use of GNN's for community detection in Parkinson's Disease (PD). Currently PD individuals are mostly classified whether they have a genetic mutation (family history) or if the onset of their disease is considered idiopathic (sporadic). Using multiple sources of data to cluster individuals with PD could result in novel understandings and relationships in PD. A sample use case of this work could be in the use of targeted clinical trials.
Parkinson’s Progressive Marker Inititative (PPMI)
 PPMI was designed with the intention of identifying PD progression biomarkers and to subset individuals with PD by their clinical, genetic and biomarker signatures. The PPMI dataset is an observational,
international, multicenter longitudinal study of individuals with PD, individuals at risk
of PD and healthy volunteers (Craig et al. 2021). Participants with PD in this study had a clinical diagnosis of PD for less than 2 years, were not taking PD medication before enrolment
and demonstrated a dopaminergic deficit determined by a visual dopamine transporter
(DaT) scan evaluation at the time of enrolment (Craig et al. 2021). Genetic risk loci have been discovered though genome-wide association studies (Bandres-Ciga et al. 2020). Genes discovered include but are
not limited to SNCA, LRRK2 and GBA. The PPMI dataset has labelled individuals at
a higher than normal risk of PD as having a mutation in one of theses genes. The PPMI study has collected longitudinal data consisting of genetic, image, lifestyle and other data types.
PPMI was designed with the intention of identifying PD progression biomarkers and to subset individuals with PD by their clinical, genetic and biomarker signatures. The PPMI dataset is an observational,
international, multicenter longitudinal study of individuals with PD, individuals at risk
of PD and healthy volunteers (Craig et al. 2021). Participants with PD in this study had a clinical diagnosis of PD for less than 2 years, were not taking PD medication before enrolment
and demonstrated a dopaminergic deficit determined by a visual dopamine transporter
(DaT) scan evaluation at the time of enrolment (Craig et al. 2021). Genetic risk loci have been discovered though genome-wide association studies (Bandres-Ciga et al. 2020). Genes discovered include but are
not limited to SNCA, LRRK2 and GBA. The PPMI dataset has labelled individuals at
a higher than normal risk of PD as having a mutation in one of theses genes. The PPMI study has collected longitudinal data consisting of genetic, image, lifestyle and other data types.
References
Hauser, Robert A. (2010). “Early Pharmacologic Treatment in Parkinson’s Disease”. In: Supplements and Featured Publications 16 (4). https://www.ajmc.com/view/a280_10mar_hauser.
Klein, Christine and Ana Westenberger (Jan. 2012). “Genetics of Parkinson’s Disease”. In: Cold Spring Harbor Perspectives in Medicine 2 (1), a008888. doi: 10.1101/CSHPERSPECT.A008888.
Dextera, David T. and Peter Jenner (2013). “Parkinson disease: From pathology to molec- ular disease mechanisms”. In: Free Radical Biology and Medicine 62, pp. 132–144. issn: 18734596. doi: 10.1016/J.FREERADBIOMED.2013.01.018.
Pai, Shraddha and Gary D. Bader (Sept. 2018). “Patient Similarity Networks for Precision Medicine”. In: Journal of molecular biology 430 (18 Pt A), pp. 2924–2938. issn: 1089-8638. doi: 10.1016/J.JMB.2018.05.037.
David W. Craig, Elizabeth Hutchins, Ivo Violich, Eric Alsop, J. Raphael Gibbs, Shawn Levy, Madison Robison, Nripesh Prasad, Tatiana Foroud, Karen L. Crawford, Arthur W. Toga, Timothy G. Whitsett, Seungchan Kim, Bradford Casey, Alyssa Reimer, Samantha J. Hutten, Mark Frasier, Fabian Kern, Tobias Fehlman, Andreas Keller, Mark R. Cookson, Kendall Van Keuren-Jensen, Samantha Hutten, and Kendall Van Keuren-Jensen. RNA sequencing of whole blood reveals early alterations in immune cells and gene expression in parkinson’s disease. Nature Aging 2021 1,734–747. doi: 10.1038/s43587-021-00088-6.
Sara Bandres-Ciga, Monica Diez-Fairen, Jonggeol Jeff Kim, and Andrew B. Singleton. Genetics of parkinson’s disease: An introspection of its journey towards precision medicine. Neurobiology of Disease, 137:104782, 4 2020. doi: 10.1016/j.nbd.2020.104782.